Click here for Part 2, The Literature Review.
A gene promoter is a region of DNA which acts like a switch to “turn a gene on”. The more methylated the promoter is, the more it is “turned on”.
RNA is a key molecule found in cells responsible for a variety of biochemical processes that are essential to the integrity of the cell (D’Aquila, 2017). Hypomethylation (reduced levels of methylation) of the RNA gene promoter has been observed in many different types of cancer (Ghoshal K, 2004). Additionally, hypermethylation (high levels of methylation) of the RNA promoter gene has also been associated with Alzheimer’s disease (Pietrzak M, 2011).
Within ageing research, there is growing interest in the RNA promoter and it’s methylation status. This is because of it’s association with age related disease (D’Aquila, 2017).
What Is DNA methylation?
DNA Methylation is used to control gene expression and maintain stability in the genome. Methylation refers to the addition of a methyl group via a covalent bond at the fifth carbon on a cytosine base within a CpG dinucleotide giving rise to 5-methyl cytosine (Jin, 2011). A CpG dinucleotide is a site where a cytosine base lies next to a Guanine base in the DNA sequence connected via a phosphodiester bond (Jr, 2017). Hypermethylation of CpG sites is often associated with transcriptional repression, conversely, hypomethylation is associated with transcriptional activation (Bird, 1992). DNA methylation prevents the binding of transcription factors to the DNA or leading to transcriptional silencing (Bird, 2001). Special enzymatic molecules called DNA Methyltransferases (DMNT1, DMNT3a and DMNT3b) catalyse DNA methylation. This is outlined in figure 1.1 (Cheng, 2008).
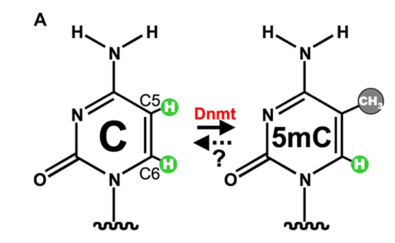
Figure 1.1: DNA Cytosine ring showing the process in which the DNA methyltransferase enzyme (DNMT) facilitates methylation on the Carbon 5 position. Figure Taken from (Cheng, 2008).
Why Is DNA Methylation So Important?
DNA methylation occurs throughout life and is essential for normal development beginning during embryonic development. DNA methylation is carried out in two different forms. Maintenance methylation and de novo methylation. The most common enzyme within maintenance methylation is DNA methyltransferase 1 (DNMT1) which is used to methylate hemi-methylated CpG dinucleotides in the genome, ensuring reformation of parental DNA methylation pattern which can be lost in the daughter DNA (Chen T, 2006).Within de novo methylation however, the DNMT3 family of catalytic enzymes are present. These are DNMT3a and DNMT3b which can newly methylate cytosine groups. Predominantly occurring within the embryo development stages of a mammal’s life cycle. There is also another enzyme present within the DNMT3 family known as DNMT3L which is largely inactive but has been observed to stimulate methylation of DNA by DNMT3a when they are both co-expressed (D’Aquila, 2017). The difference between de novo and maintenance methylation is illustrated below in figure 1.2.
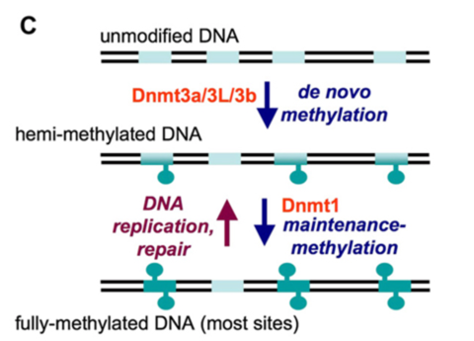
Figure 1.2: De novo methylation vs maintenance methylation. The pale blue segments illustrate substrate sequences (mainly CpG sites) whilst the turquoise shapes represent methyl groups on cytosines. After replication or repair the duplex is methylated on a single strand only. Figure taken from (Cheng, 2008).
References
Åsa Johansson, S. E. (2013). Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan. PLOS ONE, e67378.
Andrew E. Teschendorff, U. M.-M. (2010). Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Research, 440-446.
Armstrong NJ, M. K. (2017). Aging, exceptional longevity and comparisons of the Hannum and Horvath epigenetic clocks. Epigenomics, 9, 689-700.
Bartlett, Z. (2014, November 14). The Hayflick Limit. Retrieved from The Embryo Project Encyclopedia: https://embryo.asu.edu/pages/hayflick-limit
Berdyshev GD, K. G. (1967). Nucleotide composition of DNA and RNA from somatic tissues of humpback and its changes during spawning. Biokhimiya, 32, 988-993.
Bilian Jin, Y. L. (2011). DNA Methylation: Superior or Subordinate in the Epigenetic Hierarchy? Genes & Cancer, 607–617.
Bird. (1992). The essentials of DNA methylation. Cell 70, 5-8.
Bird, A. (2001). Methylation talk between histones and DNA. Science , 2113–2115.
Bjornsson HT, S. M. (2008). Intra-individual change over time in DNA methylation with familial clustering. The Journal of the American Medical Association, 299, 2877-2883.
Bocklandt S, L. W. (2011). Epigenetic predictor of age. PLOS One, 6, e14821.
Casillas MA Jr, L. N. (2003). Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Molecular and Cellular Biochemistry, 252, 33-43.
Chao Sheng, J. J. (2018). A stably self-renewing adult blood-derived induced neural stem cell exhibiting patternability and epigenetic rejuvenation. Nature Communications 9, Article number: 4047.
Chen T, L. E. (2006). Establishment and maintenance of DNA methylation patterns in mammals. Current Topics in Microbiology and Immunology, 179-201.
D’Aquila, P. (2017). Methylation of the ribosomal RNA gene promoter is associated with aging and age related decline. Aging Works, 966-975.
Feinberg AP, I. R. (2010). Personalized epigenomic signatures that are stable over time and covary with body mass index. Science Translational Medicine, 2, 49ra67.
Field AE, R. N. (2018). DNA Methylation Clocks in Aging: Categories, Causes, and Consequences. Molecular Cell, 882-895.
Fraga MF, B. E. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America, 102, 10604-10609.
Ghoshal K, M. S. (2004). Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. Journal of Biological Chemistry, 6783–6793.
Hannum G, G. J. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell, 359-367.
Horvath S, a. R. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nature Reviews Genetics 19, 371–384.
Horvath S, P. C. (2015). Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging, 1159–70.
Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, R115.
Issa JP, O. Y. (1994). Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nature Genetics, 7, 536-540.
J.Catania, D. (1991). DNA methylation and cellular ageing. Mutation Research/DNAging, 283-293 .
Jr, W. C. (2017). Medical Definition of CpG . Retrieved from Medicine Net: https://www.medicinenet.com/script/main/art.asp?articlekey=26443
Kalyan K. Pasumarthy, N. D. (2017). Methylome Analysis of Human Bone Marrow MSCs Reveals Extensive Age- and Culture-Induced Changes at Distal Regulatory Elements. Stem Cell Reports, 999–1015.
Kananen L, M. S. (2016). The trajectory of the blood DNA methylome ageing rate is largely set before adulthood: evidence from two longitudinal studies. Age, 3, Article number: 65.
Kang GH, L. S. (2003). Profile of aberrant CpG island methylation along the multistep pathway of gastric carcinogenesis. Laboratory investigation; a journal of technical methods and pathology, 83, 635-641.
Karen A. Lillycrop, S. P. (2013). DNA methylation, ageing and the influence of early life nutrition . Proceedings of the Nutrition Society, 413–421.
Lisa M. McEwen, A. M. (2017). Differential DNA methylation and lymphocyte proportions in a Costa Rican high longevity region. Epigenetics & Chromatin, 10, Article number: 21.
Loukas Zagkos, M. M. (2019). Mathematical models of DNA methylation dynamics: Implications for health and ageing. Journal of Theoretical Biology, 184-193.
Maegawa, S. (2017). Caloric restriction delays age-related methylation drift. Nature Communications, 539.
Mendelsohn, L. (2017). Epigenetic Drift Is a Determinant of Mammalian Lifespan. Rejuvenation Research, 430-436.
Pappas, S. (2018, January 30). Weird: Naked Mole Rats Don’t Die of Old Age. Retrieved from Live Science: https://www.livescience.com/61568-naked-mole-rats-no-aging.html
Peter D. Fransquet, J. W. (2019). The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clinical Epigenetics, 11:62.
Pietrzak M, R. G. (2011). Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS ONE, e22585.
Sonia Shah, A. F. (2014). Genetic and environmental exposures constrain epigenetic drift over the human life course. Genome Research, 1725-1733.
Singhal RP, M.-H. L. (1987). DNA methylation in aging of mice. Mechanisms of Ageing and Development, 41, 199–210.
Sun, D. (2014). Epigenomic Profiling of Young and Aged HSCs Reveals Concerted Changes during Aging that Reinforce Self-Renewal. Cell Stem Cell, 673-688.
Talens RP, B. D. (2010). 45. Talens RP, Boomsma DI, Tobi EW et al. (2010) Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. The FASEB Journal, 24, 3135-3144.
Xiaodong Cheng, a. R. (2008). Mammalian DNA Methyltransferases: A Structural Perspective. Structure, 341-350.

Hassan graduated with a Master’s degree in Chemical Engineering from the University of Chester (UK). He currently works as a design engineering consultant for one of the largest engineering firms in the world along with being an associate member of the Institute of Chemical Engineers (IChemE).


